Big Data Genomics – How to efficiently store and retrieve mutation data
This blog post is the first one in a series of articles that describe the use of NoSQL databases to efficiently store and retrieve mutation data.
- 1. Part one introduces the notion of mutation data and describes the conceptual use of the Cassandra NoSQL datastore.
The only way to learn a new technology is by putting it into practice. Just try to find a suitable use case in your immediate working environment and give it go. In my case, it was trying to efficiently store and retrieve mutation data through a variety of NoSQL data stores, including Cassandra, MongoDB and Neo4J.
1. What is mutation data?
DNA, or deoxyribonucleic acid, is the hereditary material that defines an organism. DNA information is stored as a code made up of four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T). The human DNA for instance, contains 3 billion bases. The order, or sequence, of these bases defines the information available for building and maintaining an organism, similar to the way in which letters of the alphabet appear in a certain order to form words and sentences.
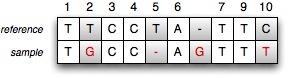
A mutation is a change in the DNA sequence of an organism. Mutations can happen for various reasons, the most common one being an error when DNA material is being copied. If the reference DNA sequence is available for a particular organism, one can try to identify the mutations between this reference and the DNA material that is extracted from a similar organism. Different types of mutations are possible. A point mutation is a mutation where one base mutated into another base. In the example below, two point mutations can be identified: the reference T base at position 2 mutated into a G base and the reference C base at position 10 mutated into a T base. At position 5, a deletion is found: the reference sequence has a T base which can not be found in the sample sequence. The inverse is also possible: the sample sequence inserts a G base at position 6.
Scientific insights can be gained by observing how organisms mutate over time. Antiviral drug research for instance, tries to identify which mutations can be candidates for applying fake DNA building blocks. If a virus can be tricked into mistakenly incorporating these fake building blocks, the effects of a virus can be reduced. An interesting question antiviral drug researchers like to see answered is the notion of mutation frequency. The mutation frequency refers to the number of times a given mutation occurs in a large population over a certain period of time. As antiviral drug researchers are typically dealing with millions of mutations, it is the ideal use case for playing around with Big Data and NoSQL.
2. Cassandra as a mutation datastore?
When working with a relational database, the first thing you do is modeling your data. A well defined database model allows you to query its data through SQL queries. Unfortunately, a fully normalized model degrades your performance when joins need to be executed on tables that contain millions of rows. To improve performance, Cassandra advocates a query-first approach, where first you identify your queries and then model your data accordingly. In the next couple of paragraphs, we will gradually explore the Cassandra data structures by developing the mutation data model. Remember, what we are trying to achieve is to be able to quickly calculate mutation frequencies!
2.1 Columns
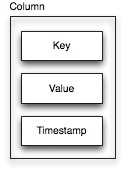
A column is Cassandra’s smallest data container. In essence, it is just a key-value pair tagged with a timestamp. (Don’t worry about the timestamp. It’s not really relevant for this discussion.)
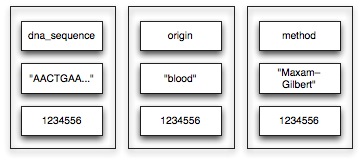
Our mutation data contains thousands of sample sequences. For each individual sequence, we would like to save several properties, including the DNA sequence, the origin and the sequencing method.
Creating columns is only the first step. Probably, you also want to group the set of columns which specify the properties of a single sequence. This grouping of columns is provided through the use of column families.
2.2 Column families
A column family is a container holding a number of rows, each row referring to a collection of columns.
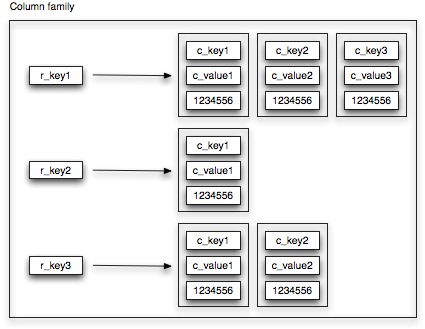
Contrary to relational database tables, column families are schema-less. When a new row is created (or updated), you can add as many columns (i.e key-value pairs) as you see fit. This allows applications to work with data in a very flexible way and enables your schema to evolve organically as new requirements pop up. Think of a column family as a HashMap of HashMaps where a unique row key refers to a set of columns, each defined by a specific column key. For each column family, you need to define how columns, contained within a row, need to be sorted. Out of the box, Cassandra provides support for BytesType, UTF8Type, LexicalUUIDType, TimeUUIDType, AsciiType, and LongType. Hence, whenever you retrieve the columns associated with a particular row, you can expect your results to be sorted as specified. If the build-in sorting types are not sufficient, you can always plug-in your own implementation.
Our mutation datastore features several column families. Let’s first have a look at our sequence column family.
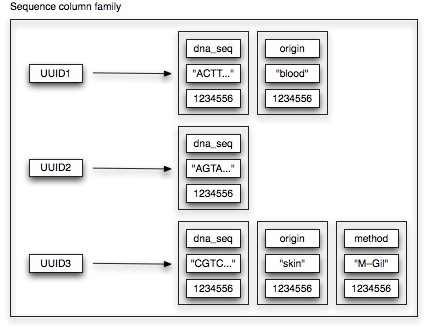
Each row needs to have an unique key. In case of the sequence column family, we use TimeUUIDs as unique keys. For each sequence, this unique TimeUUID is generated on basis of the sequence date. (Why we do this will become more clear when we talk about our mutation column family). Sorting-wise, we don’t really care how the associated columns are ordered. Hence, we just use UTF8Type sorting. As can be observed from the picture, not all properties are available for a each sequence. As no schema needs to be defined, Cassandra allows us to deal with this concept very easily.
The mutation column family features 2 Cassandra design patterns, namely Aggregate Key and Valueless Column. Remember that we want to be able to quickly retrieve all sequences that contain a particular mutation. Hence, let’s make things easy: we define the key of the mutation column family as the aggregation of all relevant mutation details (being position, type and base). If we want to retrieve all point mutations to base G at position 2, we just need to fetch the row with aggregate key 2-MUT-G. Each row refers to the list of sequences (i.e. TimeUUIDs) that contain the specific mutation. As we are basically using the row as a list, no meaningful column values are associated.
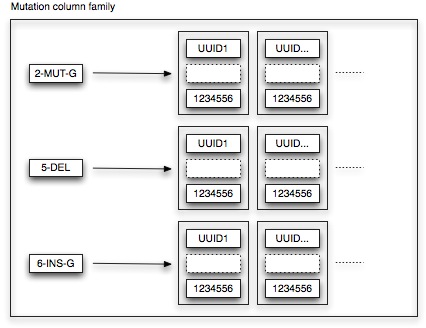
We still need to specify an ordering for the mutation column family. As the column keys are TimeUUIDs, TimeUUIDType sorting is applied. Consequently, all sequences containing the particular mutation are sorted time-wise. This design rationale allows us to fully leverage the Cassandra platform. In order to calculate the frequency of a particular mutation during a certain time period, we just need to fetch the relevant mutation row and perform a range slice query. This range slice query takes as input a lower and upper bound value (in our case the UUID representations of the start and end date of the time period) and is able to quickly retrieve the sequences within that range.
2.3 Key spaces
A key space is Cassandra’s outermost data container. It basically combines several column families in one logical space. This is similar to a relational database schema containing multiple tables.
3. Technical implementation
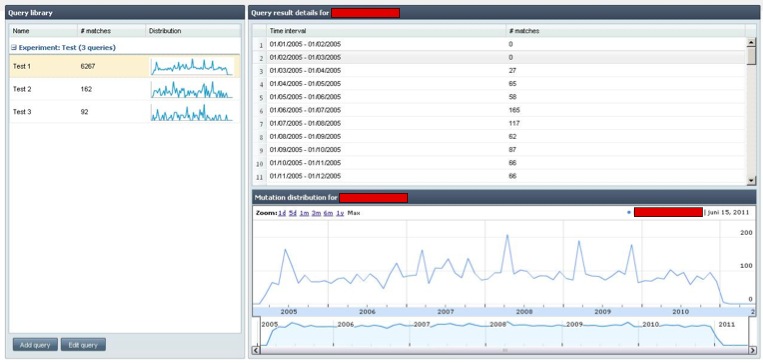
Using the concepts explained in this article, a mutation exploration tool was developed that allows scientists to query the frequency of specific mutations and compare individual results. Although the database contains millions of mutations, queries are executed blazingly fast.
4. Conclusion
This concludes the first article on saving mutation data using NoSQL datastores. The next article will provide deeper insights in how the Cassandra data model explained above is technically implemented. Looking forward to your remarks and comments!